NCERT Solutions for Class-12 Biology Chapter-11 Biotechnology: Principles and Processes
NCERT EXERCISE
Question 1. Make a chart (with diagrammatic representation) showing a restriction enzyme, the substrate DNA on which it acts, the site at which it cuts DNA and the product it produces.
Solution:
Restriction enzyme - Eco RI
Source - Escherichia coli RY13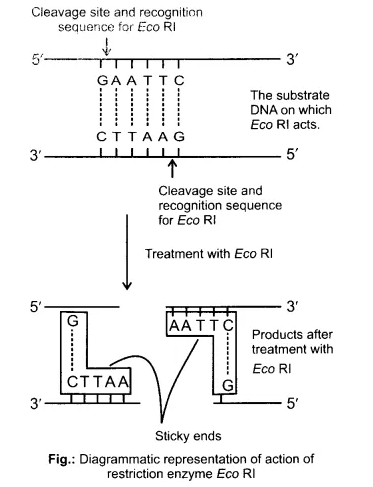
Question 2. Can you list 10 recombinant proteins which are used in medical practice? Find out where they are used as therapeutics.
Solution: The proteins produced through rDNA technology and being used in the medical practice include rh-Growth Hormone, r-Human insulin, Erythropoietin, Follicle stimulating hormone, Interferon, Insulin-like growth factor, Tissue Plasminogen Activator, factor VIII, DNase, the Envelope protein of hepatitis B virus.
Question 3. From what you have learned, can you tell whether enzymes are bigger or DNA is bigger in molecular size? How did you know?
Solution: Enzymes are bigger than DNA as they are proteins and proteins are macromolecules made of amino acids which are bigger than nucleotides. This can also be proved by gel electrophoresis, where denatured protein would not move but denatured DNA will move to a distance. Protein synthesis is regulated by small portions of DNA, called genes.
Question 4. What would be the molar concentration of human DNA in a human cell? Consult your teacher.
Solution: The average molecular weight of a nucleotide in human DNA is 130.86. The molecular weight of human DNA will therefore be 6 x 109 nucleotides (based on the human genome project) x 130-86 = 784-56 x 109 gm/mol. The molar concentration of DNA can be calculated accordingly.
The molarity can be calculated as,
Molar Concentration =
Solution: Restriction enzymes also called ‘the molecular scissors’ are used to break DNA molecules. These enzymes are present in many bacteria where they function as a part of their defence mechanism called a restriction-modification system. The molecular basis of this system was explained first by W. Arber in 1965. The restriction-modification system consists of two components,
A restriction enzyme which identifies the introduced foreign DNA and cuts into pieces and is called restriction endonuclease,
The second component is a modification enzyme in which methylation is done. Once a base in a DNA sequence is modified by the addition of a methyl group, the restriction enzymes fail to recognise and could not cut that DNA. This is how a bacterium modifies and therefore, protects its own chromosomal DNA from cleavage by these restriction enzymes. Eukaryotic cells do not have restriction endonucleases (or restriction-modification system). The DNA molecules of eukaryotes are heavily methylated by a modification enzyme, called methylase. Eukaryotes exhibit some different mechanisms to counteract viral attacks.
Solution: Shake flask is used for growing microbes and mixing the desired materials on a small scale in the laboratory. However, the large-scale production of a desired biotechnological product requires large stirred tank bioreactors.
Besides better aeration and mixing properties, bioreactors have the following advantages:
1. It has an oxygen delivery system.
2. It has a foam control, temperature, and pH control system.
3. Small volumes of culture can be withdrawn periodically.
Solution: Palindromes in DNA are base pair sequences that are the same when read forward (left to right) or backward (right to left) from a central axis of symmetry. For example, the following sequences read the same on the two strands in the 5′ → 3′ direction as well as 3′ → 5′ direction.
1. 5′ – G – G – A – T – C – C – 3′
3′ – C – C – T – A – G – G – 5′
2. 5′ – G – A – A – T – T – C – 3′
5′ – C – T – T – A – A – G – 5′
3. 5′ – A – A – G – C – T – T – 3′
3′ – T – T – C – G – A – A – 5′
4. 5′ – G – T – C – G – A – C – 3′
3′ – C – A – G – C – T – G – 5′
5. 5′ – A – C – T – A – G – T – 3′
3′ – T – G – A – T – C – A – 5′
Question 8. Can you recall meiosis and indicate at what stage recombinant DNA is made?
Solution: A recombinant DNA is made in the pachytene stage of prophase I by crossing over during meiosis cell division. Recombination nodules are visible in a synaptonemal complex in the pachytene sub-stage. Crossing over occurs in this time between chromatids than recombinant DNA is formed.
Solution: Transformation is a process through which a piece of DNA is introduced into a host bacterial cell. Normally, the genes encoding resistance to antibiotics such as ampicillin, tetracycline, etc., are considered useful selectable markers to differentiate between transformed and non-transformed bacterial cell. In addition to these selectable markers, an alternative selectable marker has been developed to differentiate transformed and non-transformed bacterial cell on the basis of their ability to produce colour in the presence of a chromogenic substance.
A recombinant DNA is inserted in the coding sequence of an enzyme (5-galactosidase (reporter enzyme). If the plasmid in the bacterium does not have an insert, the presence of a chromogenic substance gives blue coloured colonies, presence of insert results into insertional inactivation of (3-galactosidase and, therefore, the colonies do not produce any colour, these colonies are marked as transformed colonies.
(a) Origin of replication
(b) Bioreactors
(c) Downstream processing
Solution:
(a) Origin of Replication (Ori) - It is a DNA sequence which is specialised to initiate replication Bacterial chromosomes and plasmids possess a single origin of replication Eukaryote chromosomes to have a number of origin of replication. Replication proceeds bidirectionally from the site of origin of replication. The sequence also possesses nearby replication control which determines the number of copies it would form. Therefore the selected plasmid should have an origin of replication that supports a high copy number.
(b) Bio-Reactor - Bio reactor used in biotechnology is generally 100-1000 litre cylindrical metal. container with a curved base to facilitate mixing of contents. The culture medium containing all nutrients, salts vitamins, hormones etc. is added along with the inoculum of transformed cells with recombinant DNA. A stirrer helps in mixing and optimum availability of nutrients to culture cells. The supply of oxygen is maintained if the cells function better under aerobic conditions. Foam is kept under control. Gadgets are attached for knowing the temperature and pH of the contents. Corrections are made when required. There is a sampling port where a small volume of culture can be withdrawn to know the growth of cells and concentration of the extractable product.
(c) Downstream processing - It is the recovery of product from fully grown genetically modified cells, its purification, and preservation. It is carried out after the sampling report indicates the completion of the biosynthetic phase and the presence of the optimum product in the cells. After leaving a part of the cellular mass of inoculum, the rest is crushed and chemically treated to separate the product. The separated product is purified and then formulated with suitable preservatives. Clinical traits are carried out to know its use and any immediate or long-term adverse effect. Every batch of the product has to pass through strict quality control testing. Of course, the procedure and vigour of downstream processing and quality control vary from product to product.
(a) PCR
(b) Restriction enzymes and DNA
(c) Chitinase
Solution:
(a) Polymerase chain reaction (PCR) is a technique of synthesizing multiple copies of the desired gene (DNA) in vitro. This technique was developed by Kary Mullis in 1985. It is based on the principle that a DNA molecule, when subjected to high temperature, splits into two strands due to denaturation. These single-stranded molecules are then converted to double-stranded molecules by synthesising new strands in presence of enzyme DNA polymerase. Thus, multiple copies of the original DNA sequence can be generated by repeating the process several times. The basic requirements of PCR are, DNA template, two nucleotide primers usually 20 nucleotides long, and enzyme DNA polymerase which is stable at high temperature (usually Taq polymerase):
The working mechanism of PCR is as follows:
First of all, the target DNA (DNA segment to be amplified) is heated to high temperature (94 to 96° C). Heating results in the separation of two strands of DNA. Each of the two strands of the target DNA now acts as a template for the synthesis of a new DNA strand. This step is called denaturation.
Denaturation is followed by annealing. During this step, two oligonucleotide primers hybridise to each single-stranded template DNA in presence of excess synthetic oligonucleotides. Annealing is carried out at lower temperatures (40° – 60°C).
The third and final step is an extension. During this step, the enzyme DNA polymerase synthesizes the DNA segment between the primers. Usually, Taq DNA polymerase, isolated from a thermophilic bacterium Thermus aquatics, is used in most cases. The two primers extend towards each other in order to copy the DNA segment lying between the two primers. This step requires the presence of deoxynucleoside triphosphates (dNTPs) and Mg2+ and occurs at 72°C.
The above-mentioned three steps complete the first cycle of PCR. The second cycle begins with denaturation of the extension product of the first cycle and after completing the extension step, two cycles are completed. If these cycles are repeated many times, the DNA segment can be amplified approximately a billion times, i.e., one billion copies of desired DNA segment are made.
(b) Restriction enzymes are used to break DNA molecules. They belong to a larger class of enzymes called nucleases. Restriction enzymes are of three types – exonucleases, endonucleases, and restriction endonucleases.
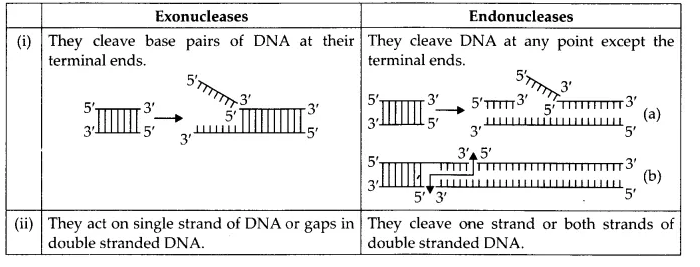
Exonucleases: They remove nucleotides from the terminal ends (either 5′ or 3′) of DNA in one strand of the duplex.
Endonucleases: They make cuts at specific positions within the DNA. These enzymes do not cleave the ends and involve only one strand of the DNA duplex.
Restriction endonucleases: These were found by Arber in 1963 in bacteria. They act as “molecular scissors” or chemical scalpels. They recognise the base sequence at palindrome sites in the DNA duplex and cut its strands. Three main types of restriction endonucleases are type I, type II, and Type III. Out of the three types, only type II restriction enzymes are used in recombinant DNA technology because they can be used in vitro to recognise and cut within specific DNA sequences typically consisting of 4 to 8 nucleotides.
(c) Chitinase is a lysing enzyme that dissolves the fungal cell wall. It results in the release of DNA along with several other macromolecules.
(a) Plasmid DNA and chromosomal DNA
(b) Exonuclease and endonuclease
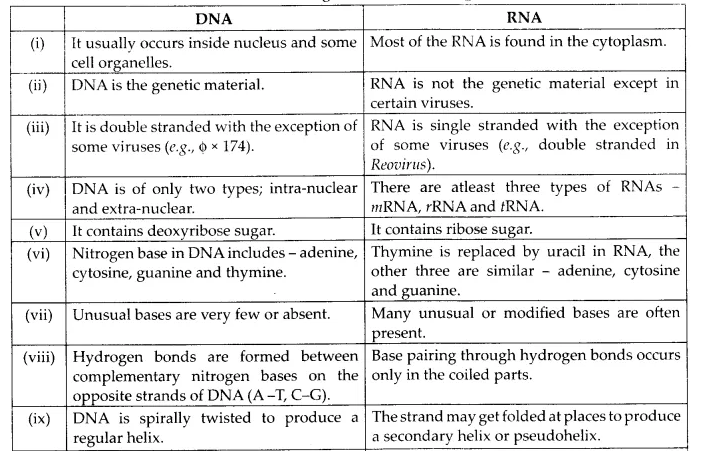
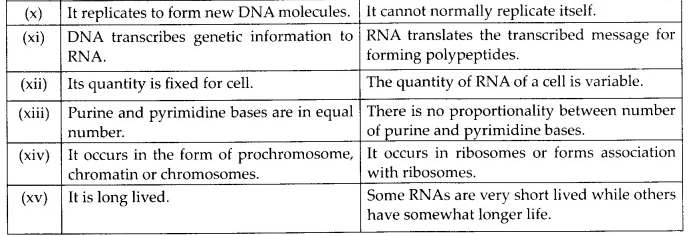
(c) RNA and DNA
Solution:
(a) Plasmid DNA is naked double-stranded DNA that forms a circle with no free ends. It is associated with few proteins. It is smaller than the host chromosome and can be easily separated.
Chromosomal DNA is a double-stranded linear DNA molecule associated with large proteins. This DNA exists in relaxed and supercoiled forms and provides a template for replication and transcription. It has free ends.
(b) Differences between exonucleases and endonucleases are as follows :
(c) Differences between DNA and RNA are given in the following table:

0 comment
Post a Comment